[1] 4 8 16Data Wrangling
Part I
ISI-BUDS 2025
Three solutions to a single problem
What is the average of 4, 8, 16 approximately?
1.What is the average of 4, 8, 16 approximately?
2.What is the average of 4, 8, 16 approximately?
3.What is the average of 4, 8, 16 approximately?
Solution 1: Functions within Functions
Problem with writing functions within functions
Things will get messy and more difficult to read and debug as we deal with more complex operations on data.
Solution 2: Creating Objects
Problem with creating many objects
We will end up with too many objects in Environment.
Solution 3: The (forward) Pipe Operator |>
Shortcut:
Ctrl (Command) + Shift + M
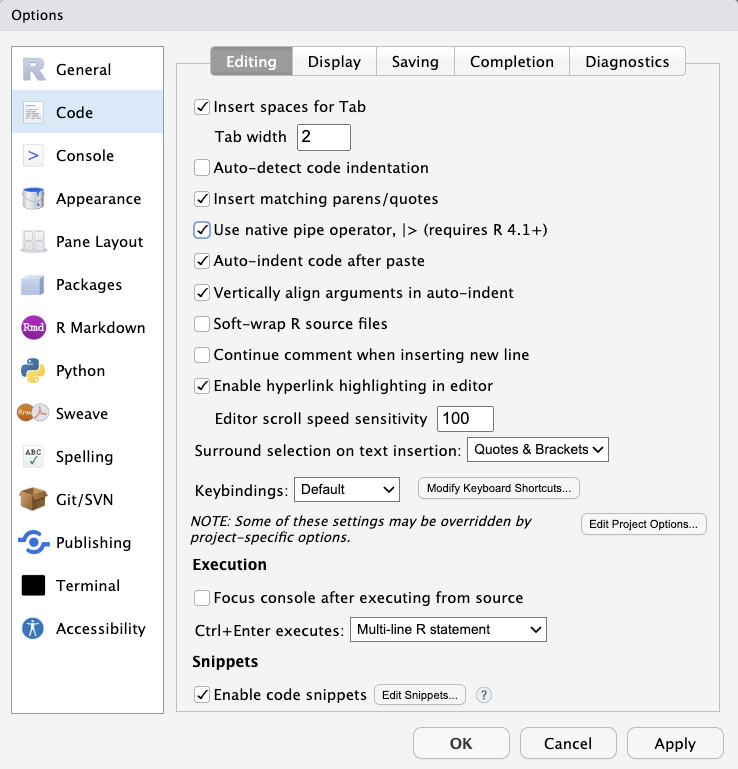
Make sure to select Use native pipe operator under Tools > Global Options > Code
Combine 4, 8, and 16 and then
Take the mean and then
Round the output
The output of the first function is the first argument of the second function.
Now we have \(f \circ g \circ h (x)\)
or round(mean(c(4, 8, 16)))
Data
Rows: 15,524
Columns: 17
$ subject_id <dbl> 1412, 533, 9134, 8518, 8967, 11048, 663, 2158, 3794, 4…
$ fake_first_name <chr> "jhezane", "penny", "grunt", "melisandre", "rolley", "…
$ fake_last_name <chr> "westerling", "targaryen", "rivers", "swyft", "karstar…
$ gender <chr> "female", "female", "male", "female", "male", "female"…
$ pan_day <dbl> 4, 7, 7, 8, 8, 8, 9, 9, 9, 9, 9, 9, 9, 9, 9, 10, 10, 1…
$ test_id <chr> "covid", "covid", "covid", "covid", "covid", "covid", …
$ clinic_name <chr> "inpatient ward a", "clinical lab", "clinical lab", "c…
$ result <chr> "negative", "negative", "negative", "negative", "negat…
$ demo_group <chr> "patient", "patient", "patient", "patient", "patient",…
$ age <dbl> 0.0, 0.0, 0.8, 0.8, 0.8, 0.8, 0.8, 0.0, 0.0, 0.9, 0.9,…
$ drive_thru_ind <dbl> 0, 1, 1, 1, 0, 0, 1, 0, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, …
$ ct_result <dbl> 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45…
$ orderset <dbl> 0, 0, 1, 1, 1, 0, 1, 1, 1, 1, 0, 1, 0, 1, 1, 1, 1, 1, …
$ payor_group <chr> "government", "commercial", NA, NA, "government", "com…
$ patient_class <chr> "inpatient", "not applicable", NA, NA, "emergency", "r…
$ col_rec_tat <dbl> 1.4, 2.3, 7.3, 5.8, 1.2, 1.4, 2.6, 0.7, 1.0, 7.1, 2.5,…
$ rec_ver_tat <dbl> 5.2, 5.8, 4.7, 5.0, 6.4, 7.0, 4.2, 6.3, 5.6, 7.0, 3.8,…See more information about the data here.
Subsetting Data Frames
subsetting variables/columns

Column-wise subsetting can be done using select().
subsetting observations/rows
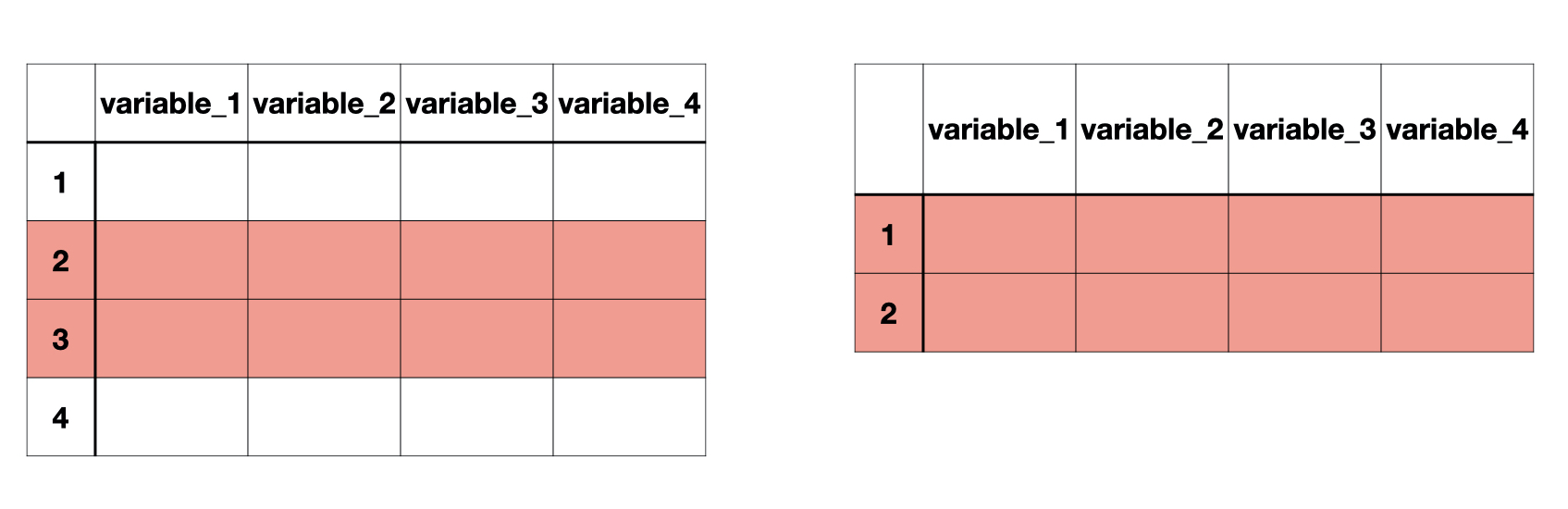
. . . Row-wise subsetting can be done with slice() and filter()
select is used to select certain variables in the data frame.
select can also be used to drop certain variables if used with a negative sign.
# A tibble: 15,524 × 15
fake_first_name fake_last_name gender pan_day test_id clinic_name result
<chr> <chr> <chr> <dbl> <chr> <chr> <chr>
1 jhezane westerling female 4 covid inpatient ward a negat…
2 penny targaryen female 7 covid clinical lab negat…
3 grunt rivers male 7 covid clinical lab negat…
4 melisandre swyft female 8 covid clinical lab negat…
5 rolley karstark male 8 covid emergency dept negat…
6 megga karstark female 8 covid oncology day ho… negat…
7 ithoke targaryen male 9 covid clinical lab negat…
8 ravella frey female 9 covid emergency dept negat…
9 styr tyrell male 9 covid clinical lab negat…
10 wynafryd seaworth male 9 covid clinical lab negat…
# ℹ 15,514 more rows
# ℹ 8 more variables: age <dbl>, drive_thru_ind <dbl>, ct_result <dbl>,
# orderset <dbl>, payor_group <chr>, patient_class <chr>, col_rec_tat <dbl>,
# rec_ver_tat <dbl>Selection helpers
starts_with()
ends_with()
contains()
# A tibble: 15,524 × 2
fake_first_name fake_last_name
<chr> <chr>
1 jhezane westerling
2 penny targaryen
3 grunt rivers
4 melisandre swyft
5 rolley karstark
6 megga karstark
7 ithoke targaryen
8 ravella frey
9 styr tyrell
10 wynafryd seaworth
# ℹ 15,514 more rows# A tibble: 15,524 × 3
fake_first_name fake_last_name clinic_name
<chr> <chr> <chr>
1 jhezane westerling inpatient ward a
2 penny targaryen clinical lab
3 grunt rivers clinical lab
4 melisandre swyft clinical lab
5 rolley karstark emergency dept
6 megga karstark oncology day hosp
7 ithoke targaryen clinical lab
8 ravella frey emergency dept
9 styr tyrell clinical lab
10 wynafryd seaworth clinical lab
# ℹ 15,514 more rowsslice() subsets rows based on a row number.
The data below include all the rows from third to seventh, including the third and the seventh.
# A tibble: 5 × 17
subject_id fake_first_name fake_last_name gender pan_day test_id clinic_name
<dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
1 9134 grunt rivers male 7 covid clinical lab
2 8518 melisandre swyft female 8 covid clinical lab
3 8967 rolley karstark male 8 covid emergency de…
4 11048 megga karstark female 8 covid oncology day…
5 663 ithoke targaryen male 9 covid clinical lab
# ℹ 10 more variables: result <chr>, demo_group <chr>, age <dbl>,
# drive_thru_ind <dbl>, ct_result <dbl>, orderset <dbl>, payor_group <chr>,
# patient_class <chr>, col_rec_tat <dbl>, rec_ver_tat <dbl>Relational Operators in R
| Operator | Description |
|---|---|
| < | Less than |
| > | Greater than |
| <= | Less than or equal to |
| >= | Greater than or equal to |
| == | Equal to |
| != | Not equal to |
Logical Operators in R
| Operator | Description |
|---|---|
| & | and |
| | | or |
filter() subsets rows based on a condition.
The data below includes rows when the clinic name is “emergency dept”
# A tibble: 3,413 × 17
subject_id fake_first_name fake_last_name gender pan_day test_id clinic_name
<dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
1 8967 rolley karstark male 8 covid emergency d…
2 2158 ravella frey female 9 covid emergency d…
3 4930 sarra frey female 10 covid emergency d…
4 2083 weasel tarly female 10 covid emergency d…
5 10468 chella mormont female 10 covid emergency d…
6 227 maege sand female 11 covid emergency d…
7 2983 ronnel snow male 11 covid emergency d…
8 6569 lanna baelish female 11 covid emergency d…
9 8165 arianne clegane female 11 covid emergency d…
10 8395 nan frey female 11 covid emergency d…
# ℹ 3,403 more rows
# ℹ 10 more variables: result <chr>, demo_group <chr>, age <dbl>,
# drive_thru_ind <dbl>, ct_result <dbl>, orderset <dbl>, payor_group <chr>,
# patient_class <chr>, col_rec_tat <dbl>, rec_ver_tat <dbl>Q. How many tests performed in the emergency department were positive?
# A tibble: 180 × 17
subject_id fake_first_name fake_last_name gender pan_day test_id clinic_name
<dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
1 902 owen seaworth male 12 covid emergency d…
2 2573 glendon lannister male 12 covid emergency d…
3 10734 jyck sand male 12 covid emergency d…
4 5023 chataya mormont female 15 covid emergency d…
5 6493 sybelle karstark female 16 covid emergency d…
6 8662 ronald manderly male 16 covid emergency d…
7 8685 black lannister male 16 covid emergency d…
8 11411 alysane baelish female 16 covid emergency d…
9 1131 ermesande clegane female 18 covid emergency d…
10 7953 anya westerling female 18 covid emergency d…
# ℹ 170 more rows
# ℹ 10 more variables: result <chr>, demo_group <chr>, age <dbl>,
# drive_thru_ind <dbl>, ct_result <dbl>, orderset <dbl>, payor_group <chr>,
# patient_class <chr>, col_rec_tat <dbl>, rec_ver_tat <dbl>Q. How many observations are available between 10th and 50th day of the pandemic?
# A tibble: 5,667 × 17
subject_id fake_first_name fake_last_name gender pan_day test_id clinic_name
<dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
1 998 harra sand female 10 covid clinical lab
2 2103 ollo snow male 10 covid clinical lab
3 2349 yezzan royce male 10 covid line clinic…
4 4930 sarra frey female 10 covid emergency d…
5 5408 alia ryswell female 10 covid clinical lab
6 2083 weasel tarly female 10 covid emergency d…
7 8031 gueren sand male 10 covid clinical lab
8 8138 frenya swyft female 10 covid clinical lab
9 9502 lorcas mormont male 10 covid clinical lab
10 10468 chella mormont female 10 covid emergency d…
# ℹ 5,657 more rows
# ℹ 10 more variables: result <chr>, demo_group <chr>, age <dbl>,
# drive_thru_ind <dbl>, ct_result <dbl>, orderset <dbl>, payor_group <chr>,
# patient_class <chr>, col_rec_tat <dbl>, rec_ver_tat <dbl>Q. How many patients 18 years and older were tested in the emergency department?
We have done all sorts of selections, slicing, filtering on covid_data but it has not changed at all. Why do you think so?
Rows: 15,524
Columns: 17
$ subject_id <dbl> 1412, 533, 9134, 8518, 8967, 11048, 663, 2158, 3794, 4…
$ fake_first_name <chr> "jhezane", "penny", "grunt", "melisandre", "rolley", "…
$ fake_last_name <chr> "westerling", "targaryen", "rivers", "swyft", "karstar…
$ gender <chr> "female", "female", "male", "female", "male", "female"…
$ pan_day <dbl> 4, 7, 7, 8, 8, 8, 9, 9, 9, 9, 9, 9, 9, 9, 9, 10, 10, 1…
$ test_id <chr> "covid", "covid", "covid", "covid", "covid", "covid", …
$ clinic_name <chr> "inpatient ward a", "clinical lab", "clinical lab", "c…
$ result <chr> "negative", "negative", "negative", "negative", "negat…
$ demo_group <chr> "patient", "patient", "patient", "patient", "patient",…
$ age <dbl> 0.0, 0.0, 0.8, 0.8, 0.8, 0.8, 0.8, 0.0, 0.0, 0.9, 0.9,…
$ drive_thru_ind <dbl> 0, 1, 1, 1, 0, 0, 1, 0, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, …
$ ct_result <dbl> 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45…
$ orderset <dbl> 0, 0, 1, 1, 1, 0, 1, 1, 1, 1, 0, 1, 0, 1, 1, 1, 1, 1, …
$ payor_group <chr> "government", "commercial", NA, NA, "government", "com…
$ patient_class <chr> "inpatient", "not applicable", NA, NA, "emergency", "r…
$ col_rec_tat <dbl> 1.4, 2.3, 7.3, 5.8, 1.2, 1.4, 2.6, 0.7, 1.0, 7.1, 2.5,…
$ rec_ver_tat <dbl> 5.2, 5.8, 4.7, 5.0, 6.4, 7.0, 4.2, 6.3, 5.6, 7.0, 3.8,…If we want to create a smaller data frame, we can always save it in a new object:
Rows: 28
Columns: 4
$ subject_id <dbl> 1412, 533, 9134, 8518, 8967, 11048, 663, 2158, 3794, 4706,…
$ pan_day <dbl> 4, 7, 7, 8, 8, 8, 9, 9, 9, 9, 9, 9, 9, 9, 9, 10, 10, 10, 1…
$ result <chr> "negative", "negative", "negative", "negative", "negative"…
$ clinic_name <chr> "inpatient ward a", "clinical lab", "clinical lab", "clini…Changing Variables
Goal:
Create a new variable called report_delay that represents the number of hours it took for the test result to be reported inside the clinical surveillance system.
# A tibble: 15,524 × 18
subject_id fake_first_name fake_last_name gender pan_day test_id clinic_name
<dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
1 1412 jhezane westerling female 4 covid inpatient w…
2 533 penny targaryen female 7 covid clinical lab
3 9134 grunt rivers male 7 covid clinical lab
4 8518 melisandre swyft female 8 covid clinical lab
5 8967 rolley karstark male 8 covid emergency d…
6 11048 megga karstark female 8 covid oncology da…
7 663 ithoke targaryen male 9 covid clinical lab
8 2158 ravella frey female 9 covid emergency d…
9 3794 styr tyrell male 9 covid clinical lab
10 4706 wynafryd seaworth male 9 covid clinical lab
# ℹ 15,514 more rows
# ℹ 11 more variables: result <chr>, demo_group <chr>, age <dbl>,
# drive_thru_ind <dbl>, ct_result <dbl>, orderset <dbl>, payor_group <chr>,
# patient_class <chr>, col_rec_tat <dbl>, rec_ver_tat <dbl>,
# report_delay <dbl>We can use pipes with ggplot too!
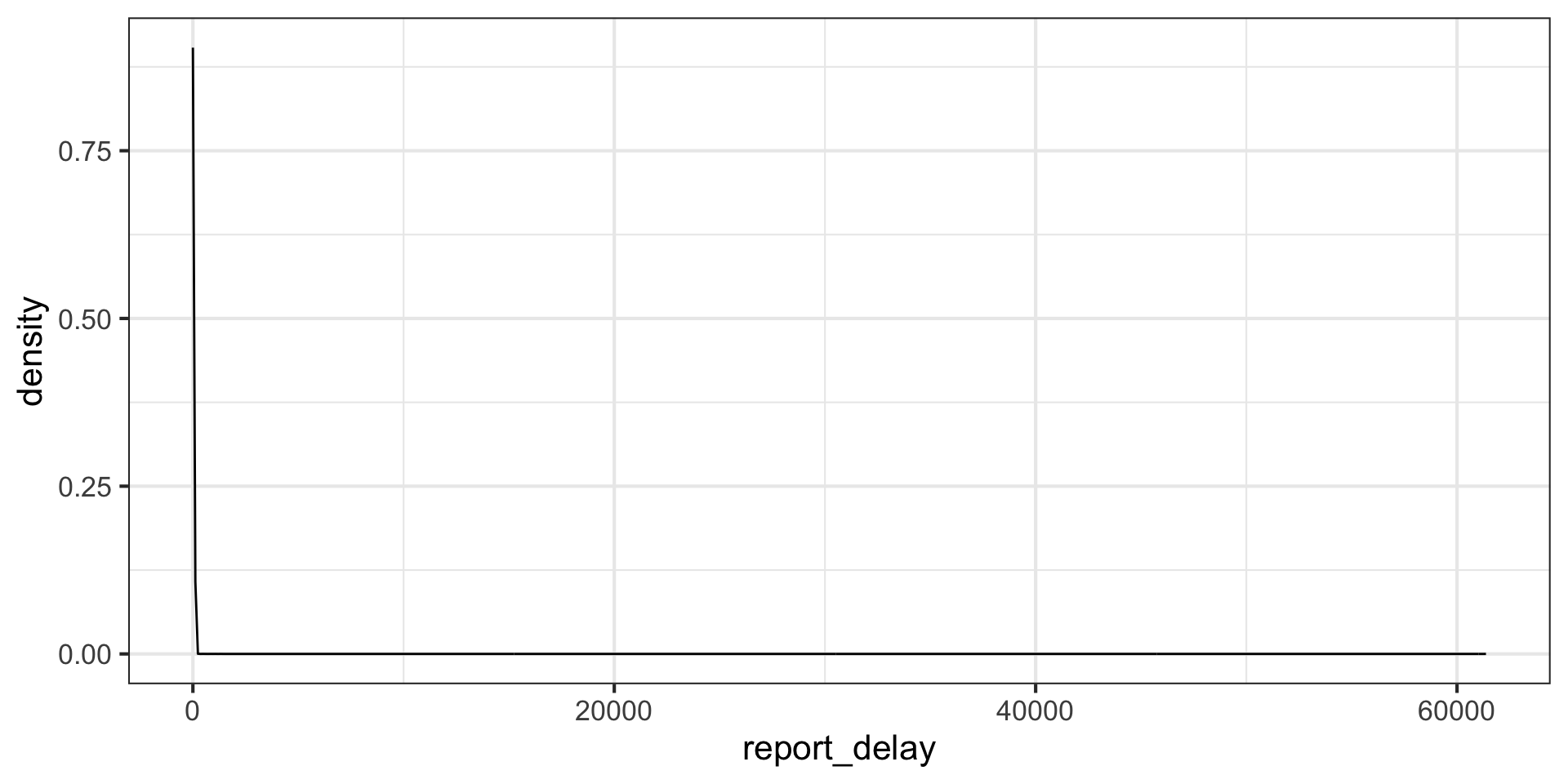
What is going on here? Maybe we have lots of outliers?
covid_data |>
mutate(report_delay = col_rec_tat + rec_ver_tat) |>
filter(report_delay > 240) # longer than 10 days# A tibble: 7 × 18
subject_id fake_first_name fake_last_name gender pan_day test_id clinic_name
<dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
1 5018 shirei royce female 41 covid inpatient wa…
2 2193 wilbert sand male 45 covid inpatient wa…
3 4860 selyse manderly female 46 covid line clinica…
4 801 tanda westerling female 56 covid 1 laboratory
5 214 rowan harlaw male 79 covid autopsy
6 1306 dick clegane male 84 covid inpatient wa…
7 11684 anguy stark male 95 covid autopsy
# ℹ 11 more variables: result <chr>, demo_group <chr>, age <dbl>,
# drive_thru_ind <dbl>, ct_result <dbl>, orderset <dbl>, payor_group <chr>,
# patient_class <chr>, col_rec_tat <dbl>, rec_ver_tat <dbl>,
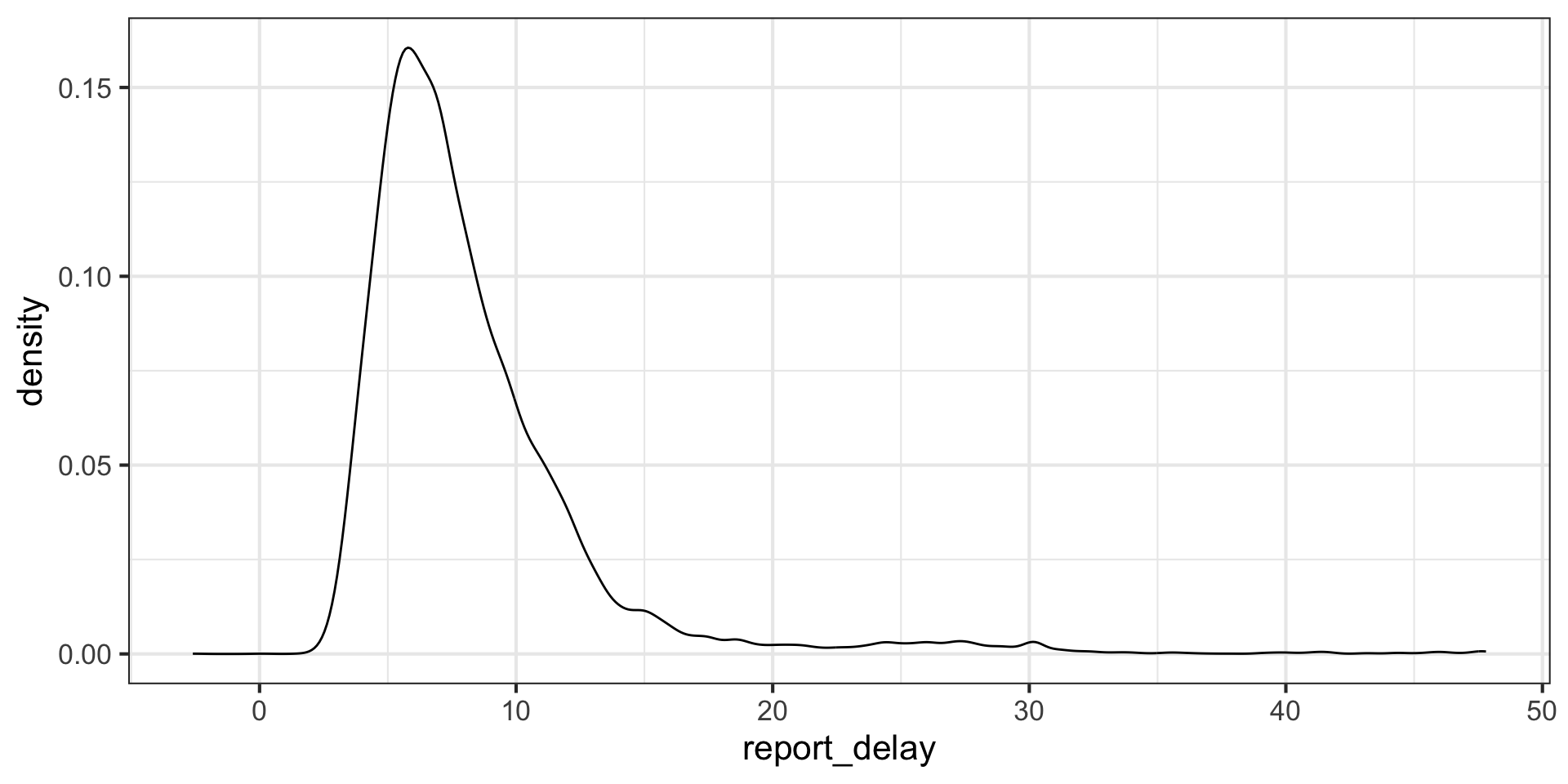
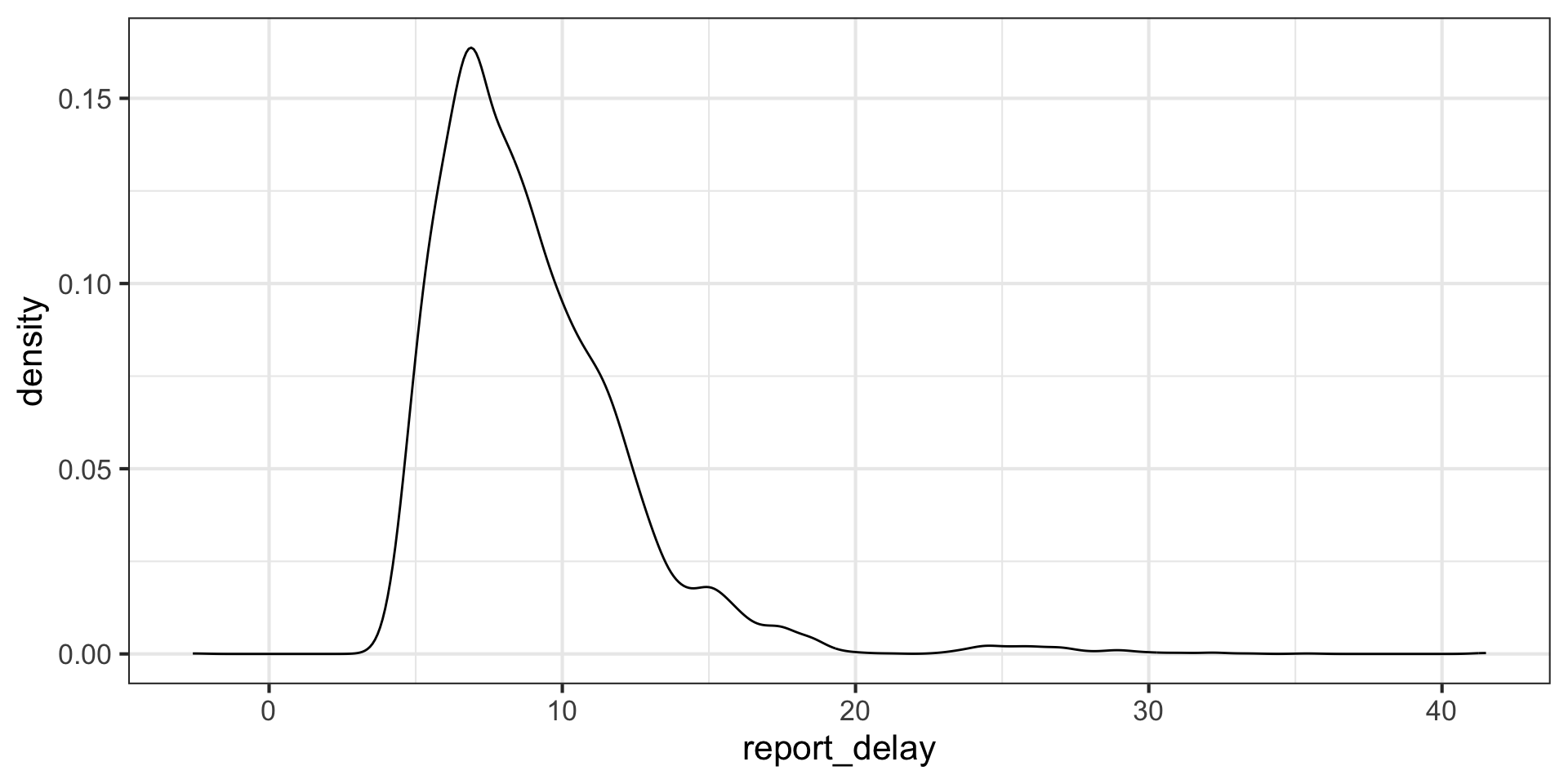
# report_delay <dbl>Let’s look at the distribution of reporting delays filtering them to be below 48 hours:
# A tibble: 102 × 4
pan_day num_positive_tests num_negative_tests total_tests
<dbl> <int> <int> <int>
1 4 0 1 1
2 7 0 2 2
3 8 0 3 3
4 9 0 9 9
5 10 1 12 13
6 11 2 69 71
7 12 4 44 48
8 13 3 28 31
9 14 2 45 47
10 15 1 46 50
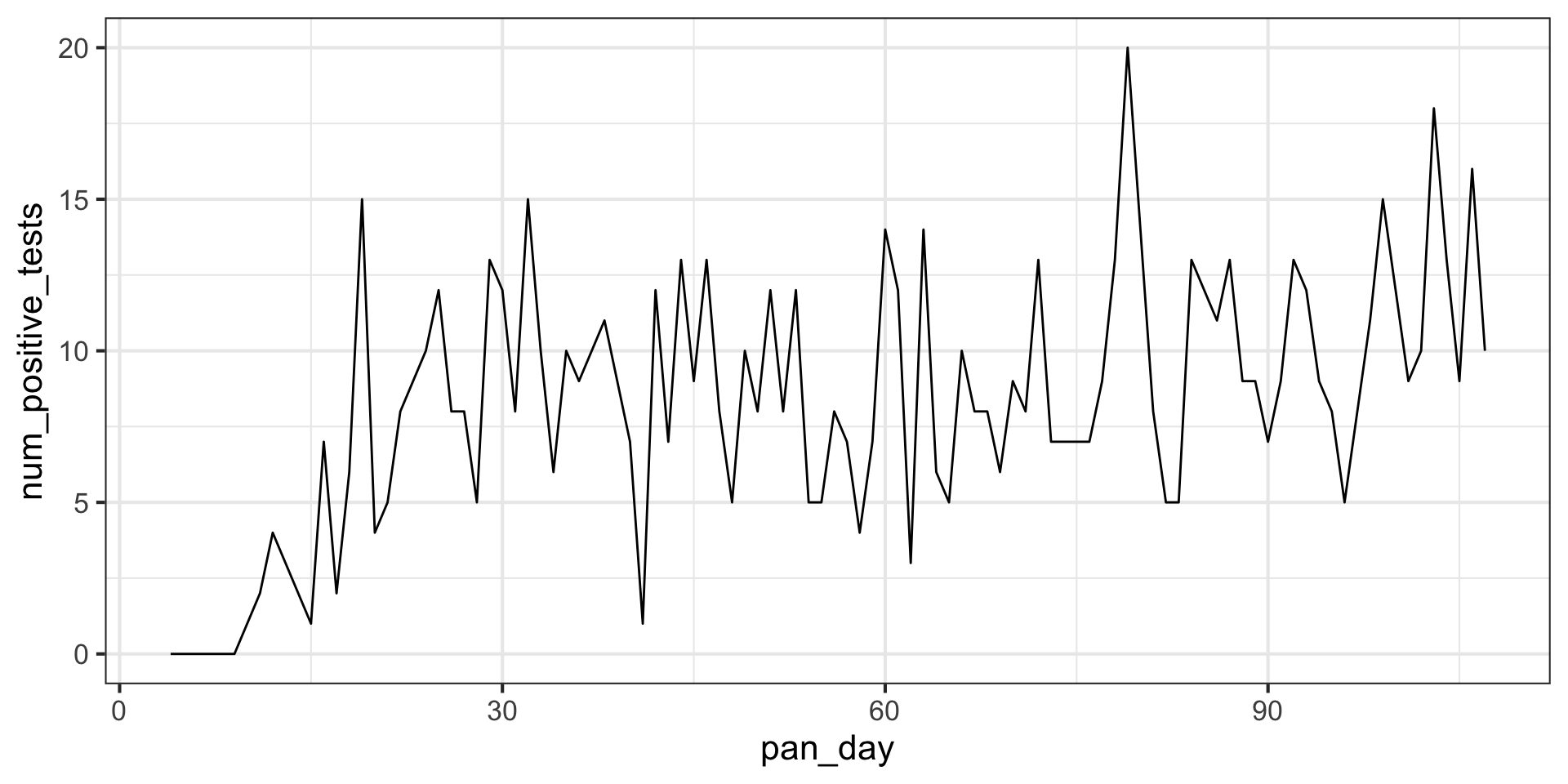
# ℹ 92 more rowsLet’s look at the positive tests as a function of time
Your task:
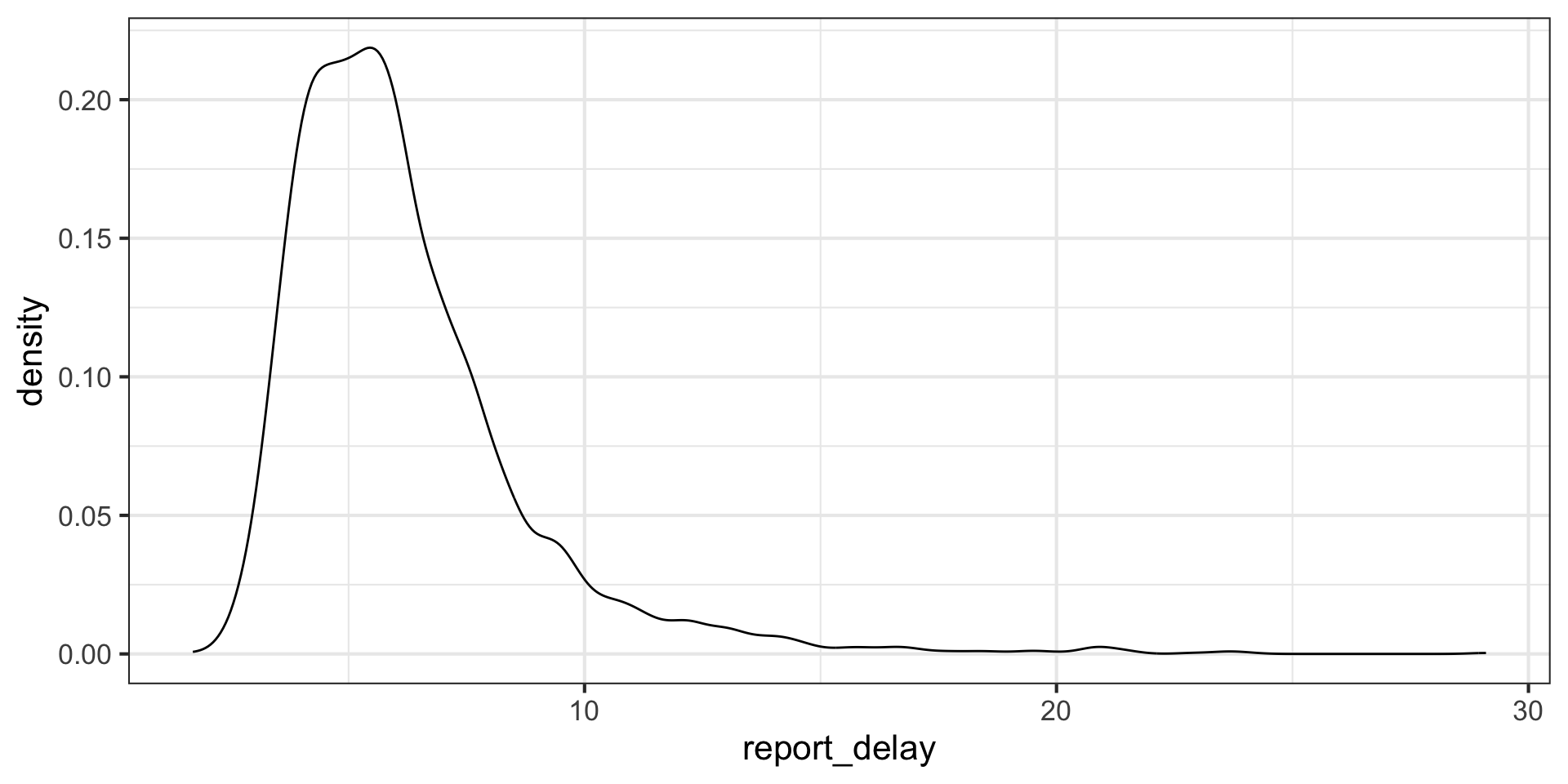
Investigate differences in reporting delays between emergency department and clinic lab tests
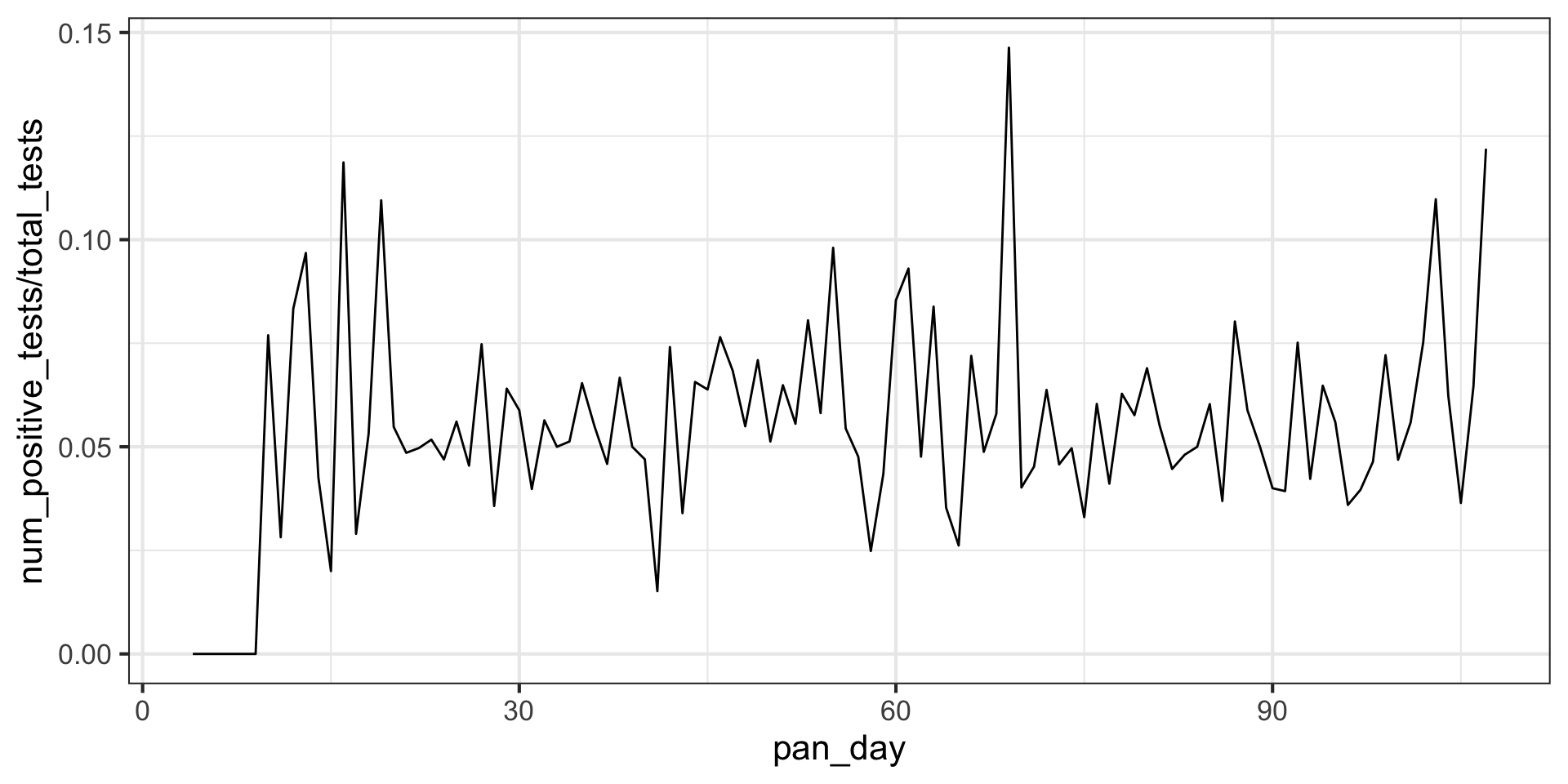
Plot test positivity (number of positive tests divided by the number of negative tests) as a function of time. What differences do you see between this time series and the time series of positive tests?
Task 1 solution:
Task 2 solution:
covid_data |>
mutate(report_delay = col_rec_tat + rec_ver_tat) |>
group_by(pan_day) |>
summarize(num_positive_tests = sum(result=="positive"), num_negative_tests = sum(result=="negative"), total_tests=n()) |>
ggplot(aes(x = pan_day, y = num_positive_tests/total_tests)) +
geom_line() +
theme_bw(base_size = 16)