Rows: 15,524
Columns: 17
$ subject_id <dbl> 1412, 533, 9134, 8518, 8967, 11048, 663, 2158, 3794, 4…
$ fake_first_name <chr> "jhezane", "penny", "grunt", "melisandre", "rolley", "…
$ fake_last_name <chr> "westerling", "targaryen", "rivers", "swyft", "karstar…
$ gender <chr> "female", "female", "male", "female", "male", "female"…
$ pan_day <dbl> 4, 7, 7, 8, 8, 8, 9, 9, 9, 9, 9, 9, 9, 9, 9, 10, 10, 1…
$ test_id <chr> "covid", "covid", "covid", "covid", "covid", "covid", …
$ clinic_name <chr> "inpatient ward a", "clinical lab", "clinical lab", "c…
$ result <chr> "negative", "negative", "negative", "negative", "negat…
$ demo_group <chr> "patient", "patient", "patient", "patient", "patient",…
$ age <dbl> 0.0, 0.0, 0.8, 0.8, 0.8, 0.8, 0.8, 0.0, 0.0, 0.9, 0.9,…
$ drive_thru_ind <dbl> 0, 1, 1, 1, 0, 0, 1, 0, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, …
$ ct_result <dbl> 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45, 45…
$ orderset <dbl> 0, 0, 1, 1, 1, 0, 1, 1, 1, 1, 0, 1, 0, 1, 1, 1, 1, 1, …
$ payor_group <chr> "government", "commercial", NA, NA, "government", "com…
$ patient_class <chr> "inpatient", "not applicable", NA, NA, "emergency", "r…
$ col_rec_tat <dbl> 1.4, 2.3, 7.3, 5.8, 1.2, 1.4, 2.6, 0.7, 1.0, 7.1, 2.5,…
$ rec_ver_tat <dbl> 5.2, 5.8, 4.7, 5.0, 6.4, 7.0, 4.2, 6.3, 5.6, 7.0, 3.8,…Data Wrangling
Part II
ISI-BUDS 2025
Data
Aggregating Data
Data
Observations
Aggregate Data
Summaries of observations
Aggregating Categorical Data
Categorical data are summarized with counts or proportions.
# A tibble: 3 × 2
result n
<chr> <int>
1 invalid 301
2 negative 14358
3 positive 865Aggregating Numerical Data
Mean, median, standard deviation, variance, and quartiles are some of the numerical summaries of numerical variables.
Aggregating Data By Groups
group_by()
We have used this function yesterday to aggregate data into daily counts of test results. Let’s review and use this function for other types of groups.

group_by() separates the data frame by the groups. Any action following group_by() will be completed for each group separately.
Q. What is the median test reporting delay for each clinic type?
# A tibble: 15,524 × 17
# Groups: clinic_name [88]
subject_id fake_first_name fake_last_name gender pan_day test_id clinic_name
<dbl> <chr> <chr> <chr> <dbl> <chr> <chr>
1 1412 jhezane westerling female 4 covid inpatient w…
2 533 penny targaryen female 7 covid clinical lab
3 9134 grunt rivers male 7 covid clinical lab
4 8518 melisandre swyft female 8 covid clinical lab
5 8967 rolley karstark male 8 covid emergency d…
6 11048 megga karstark female 8 covid oncology da…
7 663 ithoke targaryen male 9 covid clinical lab
8 2158 ravella frey female 9 covid emergency d…
9 3794 styr tyrell male 9 covid clinical lab
10 4706 wynafryd seaworth male 9 covid clinical lab
# ℹ 15,514 more rows
# ℹ 10 more variables: result <chr>, demo_group <chr>, age <dbl>,
# drive_thru_ind <dbl>, ct_result <dbl>, orderset <dbl>, payor_group <chr>,
# patient_class <chr>, col_rec_tat <dbl>, rec_ver_tat <dbl>Note that when group_by() is used there have been no changes to the number of columns or rows. The only difference we can observe is now Groups: clinic_name [88] is displayed indicating the data frame (i.e., tibble) is divided into three groups.
covid_data |>
group_by(clinic_name) |>
mutate(report_delay = col_rec_tat + rec_ver_tat) |>
filter(report_delay <= 48) |>
summarize(med_report_delay = median(report_delay))# A tibble: 88 × 2
clinic_name med_report_delay
<chr> <dbl>
1 1 laboratory 8.7
2 3 laboratory 7.05
3 anes resource ctr 5.8
4 apheresis 7.1
5 autopsy 6.7
6 bed management center 29.6
7 behavioral hosp 9.15
8 bmc 15.7
9 cardiac echo 5.6
10 cardiac ekg 5
# ℹ 78 more rowsWe can also remind ourselves how many tests were performed in each clinic group.
covid_data |>
group_by(clinic_name) |>
mutate(report_delay = col_rec_tat + rec_ver_tat) |>
filter(report_delay <= 48) |>
summarize(med_report_delay = median(report_delay), count=n())# A tibble: 88 × 3
clinic_name med_report_delay count
<chr> <dbl> <int>
1 1 laboratory 8.7 2
2 3 laboratory 7.05 2
3 anes resource ctr 5.8 3
4 apheresis 7.1 1
5 autopsy 6.7 7
6 bed management center 29.6 1
7 behavioral hosp 9.15 98
8 bmc 15.7 1
9 cardiac echo 5.6 2
10 cardiac ekg 5 7
# ℹ 78 more rowsNote that n() does not take any arguments.
Data Joins
left_join(x, y)
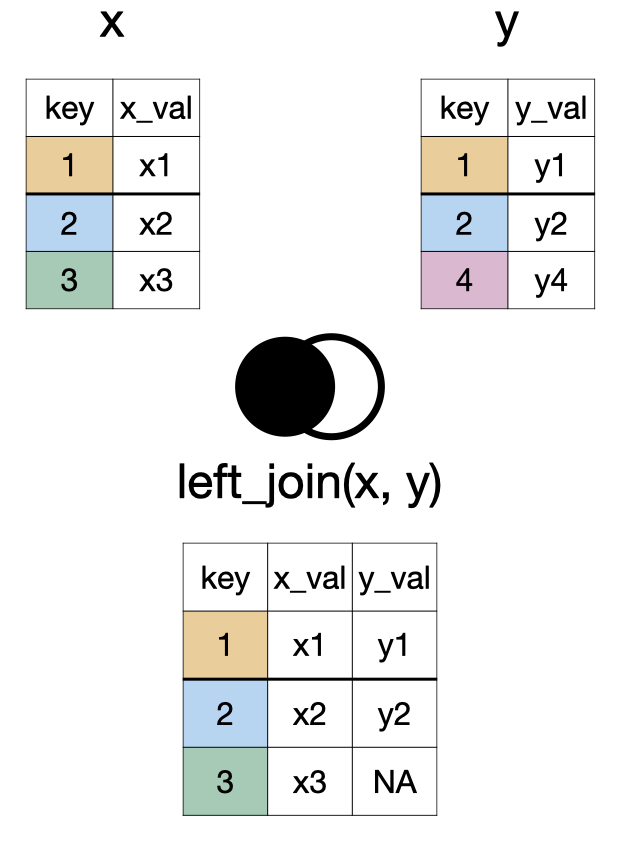
right_join(x, y)
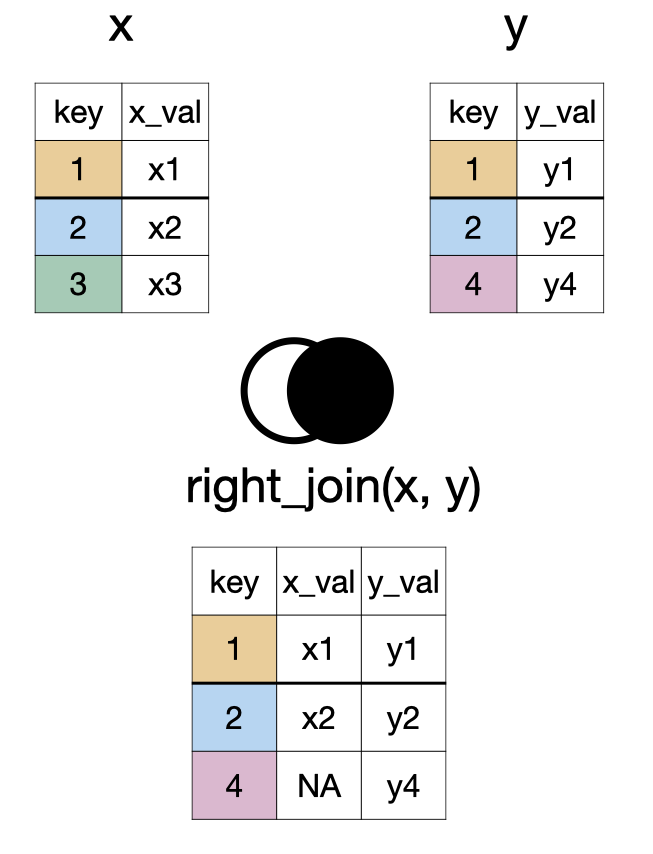
full_join(x, y)
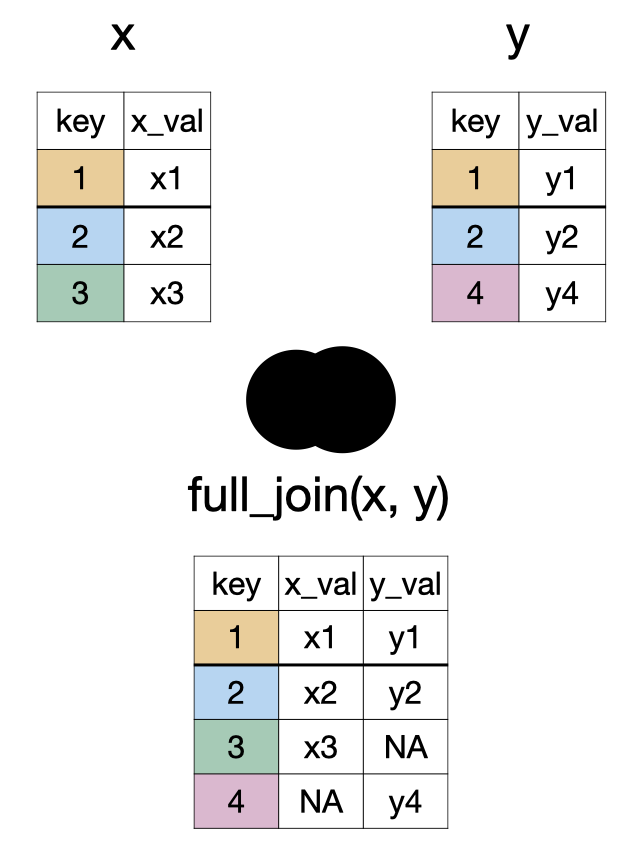
inner_join(x, y) and semi_join(x, y)
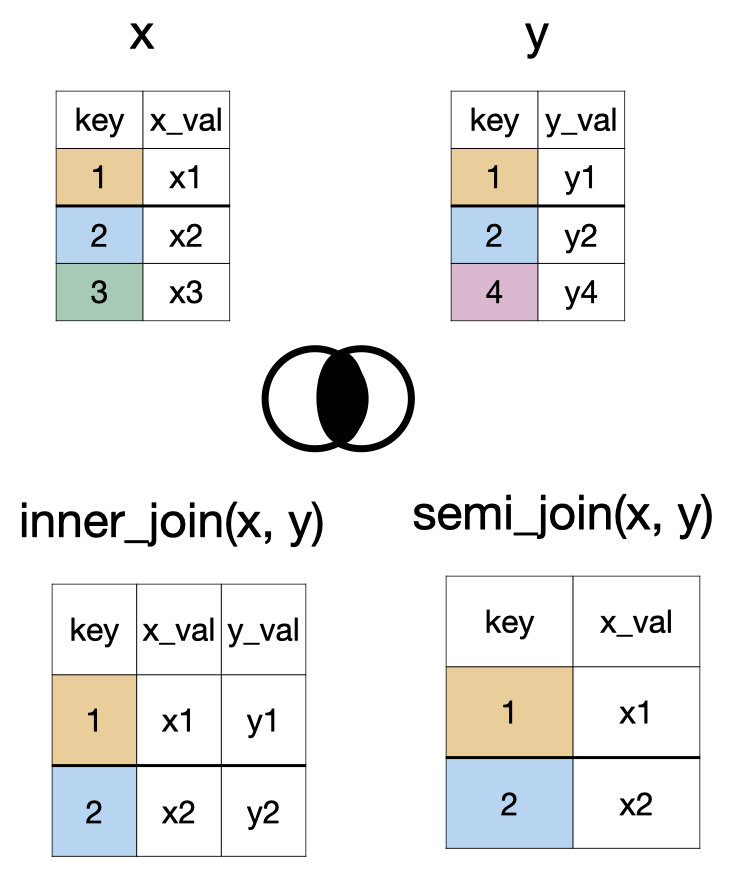
anti_join(x, y)
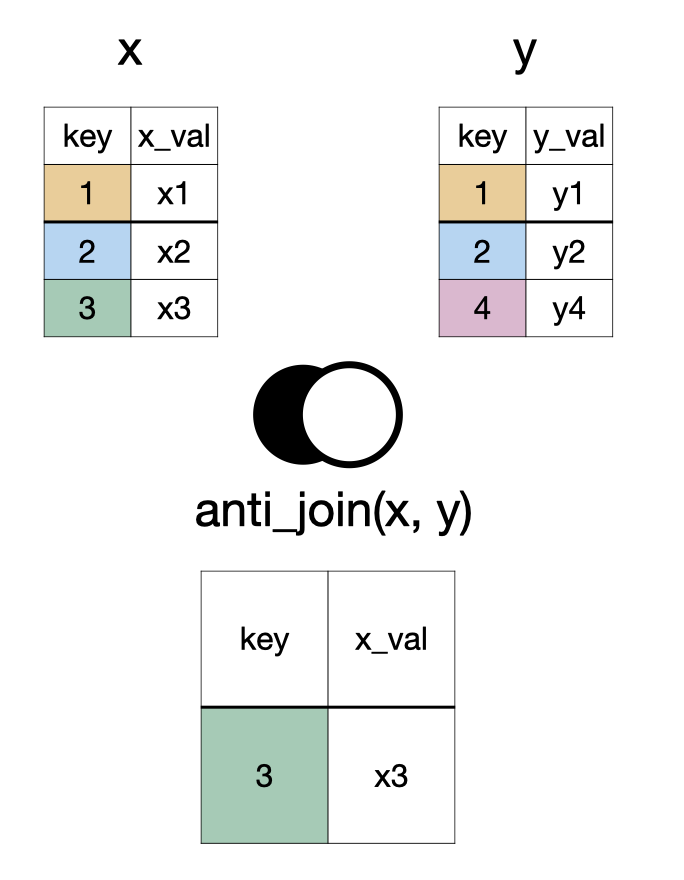
something_join(x, y)
| x | y | |||
|---|---|---|---|---|
| rows | columns | rows | columns | |
left_join()
|
all | all | matched | all |
right_join()
|
matched | all | all | all |
full_join()
|
all | all | all | all |
inner_join()
|
matched | all | matched | all |
semi_join()
|
matched | all | none | none |
anti_join()
|
unmatched | all | none | none |
# A tibble: 5 × 4
name song_name album_name song_popularity
<chr> <chr> <chr> <dbl>
1 Beyoncé Savage Remix (feat. Beyoncé) Savage Rem… 83
2 Taylor Swift cardigan folklore 85
3 Drake Laugh Now Cry Later (feat. Lil Durk) Laugh Now … 95
4 Beyoncé Halo I AM…SASHA… NA
5 Ariana Grande Stuck with U (with Justin Bieber) Stuck with… NA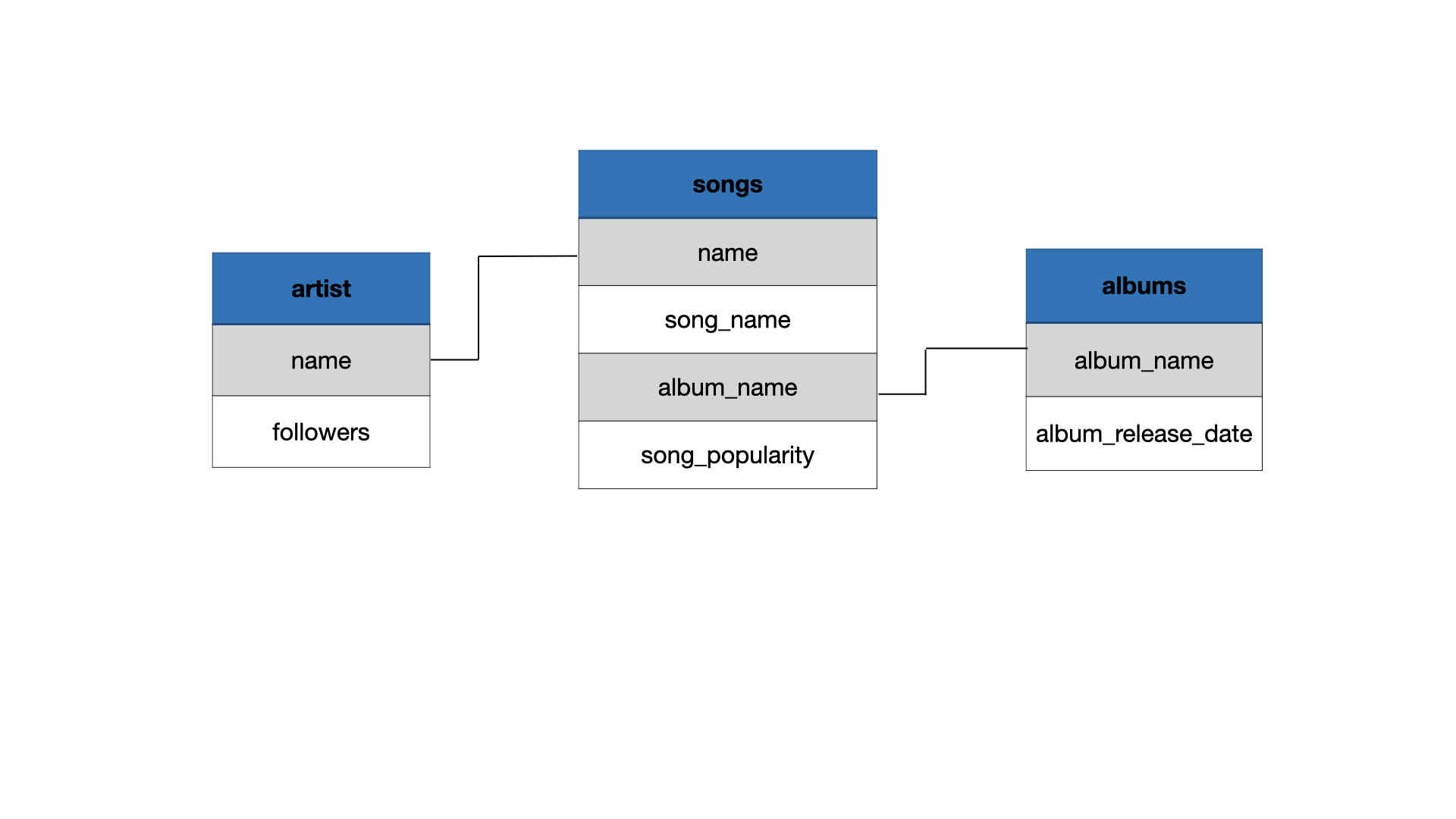
# A tibble: 5 × 5
name song_name album_name song_popularity followers
<chr> <chr> <chr> <dbl> <dbl>
1 Beyoncé Savage Remix (feat. Beyonc… Savage Re… 83 24757958
2 Taylor Swift cardigan folklore 85 33098116
3 Drake Laugh Now Cry Later (feat.… Laugh Now… 95 NA
4 Beyoncé Halo I AM…SASH… NA 24757958
5 Ariana Grande Stuck with U (with Justin … Stuck wit… NA 51807131# A tibble: 4 × 5
name song_name album_name song_popularity followers
<chr> <chr> <chr> <dbl> <dbl>
1 Beyoncé Savage Remix (feat. Beyonc… Savage Re… 83 24757958
2 Taylor Swift cardigan folklore 85 33098116
3 Beyoncé Halo I AM…SASH… NA 24757958
4 Ariana Grande Stuck with U (with Justin … Stuck wit… NA 51807131# A tibble: 5 × 5
name song_name album_name song_popularity followers
<chr> <chr> <chr> <dbl> <dbl>
1 Beyoncé Savage Remix (feat. Beyonc… Savage Re… 83 24757958
2 Taylor Swift cardigan folklore 85 33098116
3 Drake Laugh Now Cry Later (feat.… Laugh Now… 95 NA
4 Beyoncé Halo I AM…SASH… NA 24757958
5 Ariana Grande Stuck with U (with Justin … Stuck wit… NA 51807131# A tibble: 5 × 6
name song_name album_name song_popularity followers album_release_date
<chr> <chr> <chr> <dbl> <dbl> <date>
1 Beyoncé Savage R… Savage Re… 83 24757958 2020-04-29
2 Taylor Swift cardigan folklore 85 33098116 NA
3 Drake Laugh No… Laugh Now… 95 NA 2020-08-14
4 Beyoncé Halo I AM…SASH… NA 24757958 2008-11-14
5 Ariana Gran… Stuck wi… Stuck wit… NA 51807131 2020-05-08 Your tasks:
Download hospitalization data from CA Open Data Portal.
Load the data into R/RStudio using
read_csv()function and compute average number of hospital beds for each county in California over the whole time span of the data. Use hospitalized confirmed COVID patients as your variable of interest.
Your tasks:
Download a file with county population sizes from Slack.
Load the data into R/RStudio using
read_csv()function, join this table with the hospitalization data to plot a scatter plot of county averages of hospitalized COVID patients and county population sizes.Compute county average hospitalized patients per 100,000 people
ca_average_covid_hosp <- ca_covid_hosp |>
group_by(county) |>
summarize(average_hosp_covid_patients = mean(hospitalized_covid_confirmed_patients, na.rm=TRUE))
ca_average_covid_hosp# A tibble: 56 × 2
county average_hosp_covid_patients
<chr> <dbl>
1 Alameda 106.
2 Amador 3.10
3 Butte 21.0
4 Calaveras 1.11
5 Colusa 1.67
6 Contra Costa 65.7
7 Del Norte 1.87
8 El Dorado 6.04
9 Fresno 139.
10 Glenn 0.899
# ℹ 46 more rowsca_average_covid_hosp <- inner_join(ca_average_covid_hosp, ca_county_pop, by = "county")
ca_average_covid_hosp# A tibble: 56 × 3
county average_hosp_covid_patients pop_size
<chr> <dbl> <dbl>
1 Alameda 106. 1671329
2 Amador 3.10 39752
3 Butte 21.0 219186
4 Calaveras 1.11 45905
5 Colusa 1.67 21547
6 Contra Costa 65.7 1153526
7 Del Norte 1.87 27812
8 El Dorado 6.04 192843
9 Fresno 139. 999101
10 Glenn 0.899 28393
# ℹ 46 more rows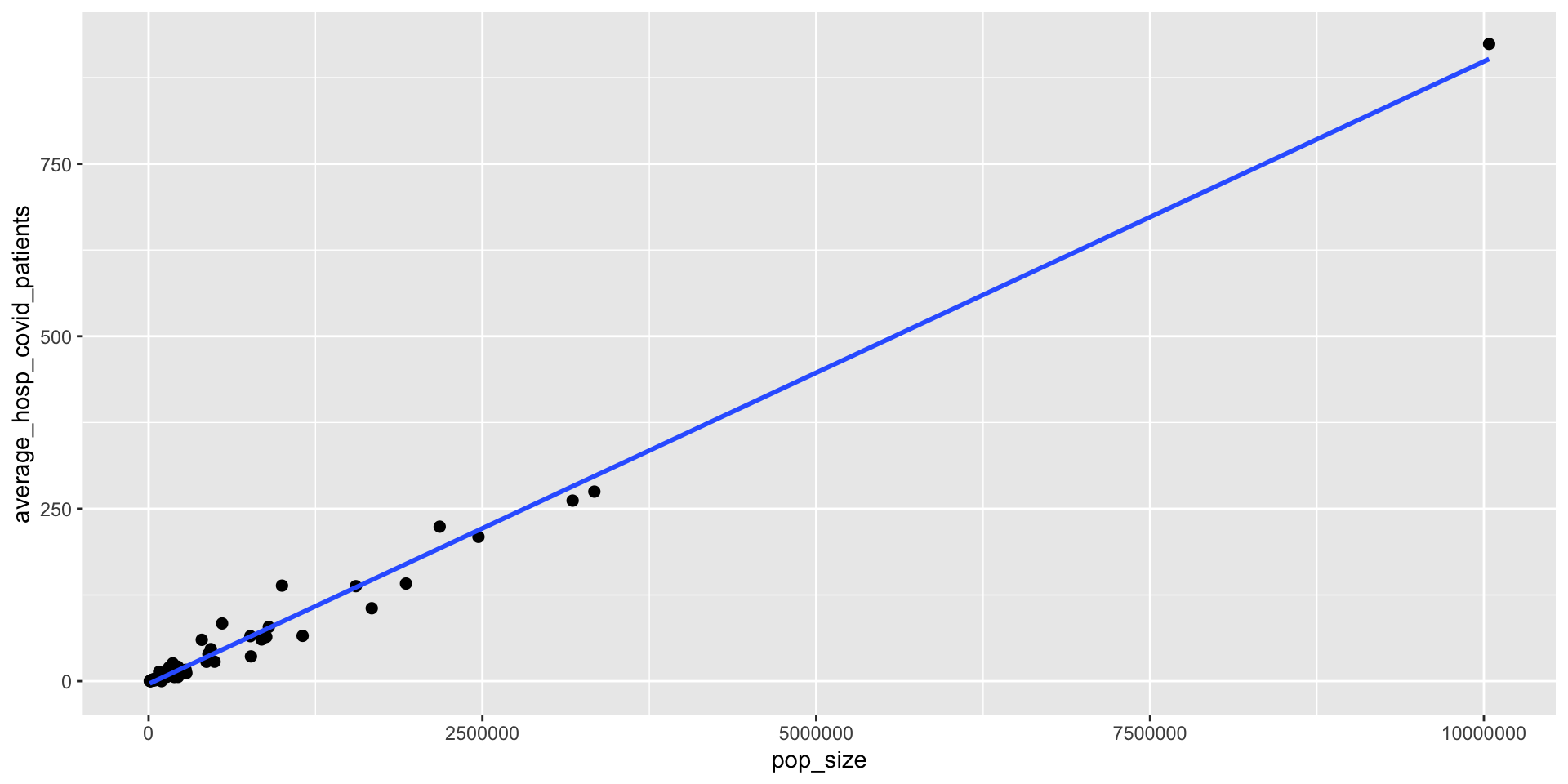
ca_average_covid_hosp |>
mutate(average_hosp_covid_patients_per_100K = average_hosp_covid_patients/pop_size*100000)# A tibble: 56 × 4
county average_hosp_covid_patients pop_size average_hosp_covid_patien…¹
<chr> <dbl> <dbl> <dbl>
1 Alameda 106. 1671329 6.33
2 Amador 3.10 39752 7.79
3 Butte 21.0 219186 9.57
4 Calaveras 1.11 45905 2.41
5 Colusa 1.67 21547 7.75
6 Contra Costa 65.7 1153526 5.70
7 Del Norte 1.87 27812 6.72
8 El Dorado 6.04 192843 3.13
9 Fresno 139. 999101 13.9
10 Glenn 0.899 28393 3.17
# ℹ 46 more rows
# ℹ abbreviated name: ¹average_hosp_covid_patients_per_100K